Supported by:
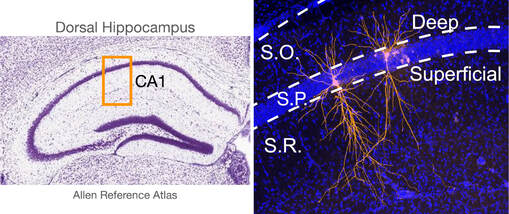
Development of neuronal subtypes and local circuits in the hippocampus
Funded by NIMH R01MH124870
Using transgenic mice, we are studying how conditional knockout of the transcription factor Satb2 alters the differentiation of deep and superficial hippocampal CA1 pyramidal cells in terms of membrane physiology, morphology, synaptic connectivity with neighboring inhibitory interneurons, and molecular expression profiled using single-cell RNA-sequencing.
Funded by NIMH R01MH124870
Using transgenic mice, we are studying how conditional knockout of the transcription factor Satb2 alters the differentiation of deep and superficial hippocampal CA1 pyramidal cells in terms of membrane physiology, morphology, synaptic connectivity with neighboring inhibitory interneurons, and molecular expression profiled using single-cell RNA-sequencing.
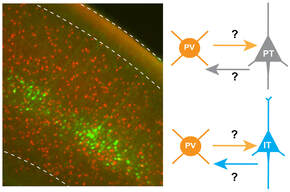
Circuit mechanisms of autism caused by embryonic Arid1b haploinsufficiency
Funded by the Simons Foundation Autism Research Initiative (SFARI)
Project site
We are using transgenic mice to study how Arid1b haploinsufficiency in different types of excitatory projection neurons or inhibitory interneurons differentially contributes to abnormal circuit development and synaptic physiology, which may lead to disrupted E/I balance and autism spectrum disorder.
Funded by the Simons Foundation Autism Research Initiative (SFARI)
Project site
We are using transgenic mice to study how Arid1b haploinsufficiency in different types of excitatory projection neurons or inhibitory interneurons differentially contributes to abnormal circuit development and synaptic physiology, which may lead to disrupted E/I balance and autism spectrum disorder.
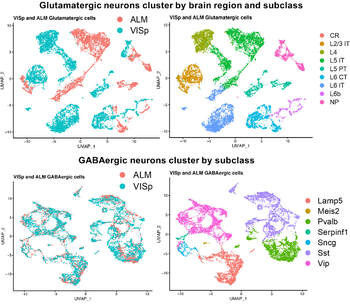
Mining the Allen Brain Institute single-cell RNA-seq database of mouse cortical neurons
We are using bioinformatic analysis of public RNA-seq datasets to compare expression of cell adhesion molecules that may be involved in the formation of local circuits among different classes of neocortical neurons both within and between distinct brain regions. We are then filtering our analyses through clinical databases to identify cell types that may play primary roles in different neurological disorders.
https://github.com/Moussa-A/Wester-Lab
We are using bioinformatic analysis of public RNA-seq datasets to compare expression of cell adhesion molecules that may be involved in the formation of local circuits among different classes of neocortical neurons both within and between distinct brain regions. We are then filtering our analyses through clinical databases to identify cell types that may play primary roles in different neurological disorders.
https://github.com/Moussa-A/Wester-Lab
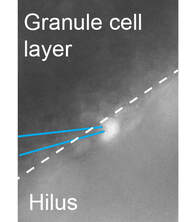
On-going collaborations:
Dr. Liz Kirby (Department of Psychology, OSU)
We are contributing physiology data to a study of how the proliferation of adult hippocampal neural stem cells is regulated by depolarization due to glutamate transport. Funded by NINDS R21NS123797 to Dr. Kirby (Co-I, Wester).
Dr. Liz Kirby (Department of Psychology, OSU)
We are contributing physiology data to a study of how the proliferation of adult hippocampal neural stem cells is regulated by depolarization due to glutamate transport. Funded by NINDS R21NS123797 to Dr. Kirby (Co-I, Wester).
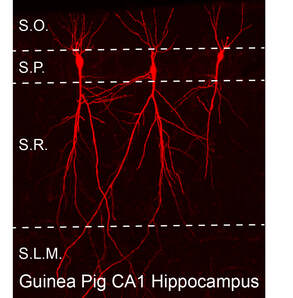
Dr. John Oberdick (Department of Neuroscience, OSU)
Single-cell RNA-seq and whole-cell patch clamp of hippocampal CA1 pyramidal cells in a guinea pig model of neonatal opioid withdrawal syndrome.
Single-cell RNA-seq and whole-cell patch clamp of hippocampal CA1 pyramidal cells in a guinea pig model of neonatal opioid withdrawal syndrome.
Dr. Paola Malerba (Nationwide Children's Hospital, Battelle Center for Mathematical Medicine)
We are contributing our physiology data from CA1 hippocampus to computational modeling of how differences between deep and superficial pyramidal cells impact the generation of network oscillations and rhythms.
We are contributing our physiology data from CA1 hippocampus to computational modeling of how differences between deep and superficial pyramidal cells impact the generation of network oscillations and rhythms.


